First-generation PPRL techniques (1998-2004) focused on matching entities that share the exact same identifying values across data sets. This was commonly done by algorithmically creating unique, encrypted, irreversible hashed pseudonyms for entities based on inputted PII, allowing data owners at different institutions to compare the one-way hashed tokens across multiple datasets and sources to match records from the same individual. A hash pseudonym can be thought of as a fingerprint of the data—it is a transformation of a given string of data into another value that represents the original string. Encryption algorithms strengthen the cybersecurity of such values because only those with a decryption key can decipher the hashed values.
Relying as they did on exact matches, these techniques could not accommodate data errors and variations, and as such, second-generation tools (2004-2009) evolved to use “fuzzy matching” to account for differences in text spelling, punctuation, and capitalization. Second-generation improvements relied on the introduction of Bloom filters, a space-efficient probabilistic data structure that is used to test whether an element is a member of a set.
As more digitized data were being collected, the third generation of PPRL techniques (2009–2014) sought to address scalability and computational resource usage.
In parallel with the big data revolution, the fourth generation of PPRL approaches (2014–2020) were tailored to optimize processes and tools for large datasets. Areas of enhancement included computational resource usage, schema optimization, privacy, and the development of tools and applications for practical utility.
More recent progress has focused on enabling use of more sophisticated analytics including machine and deep learning.

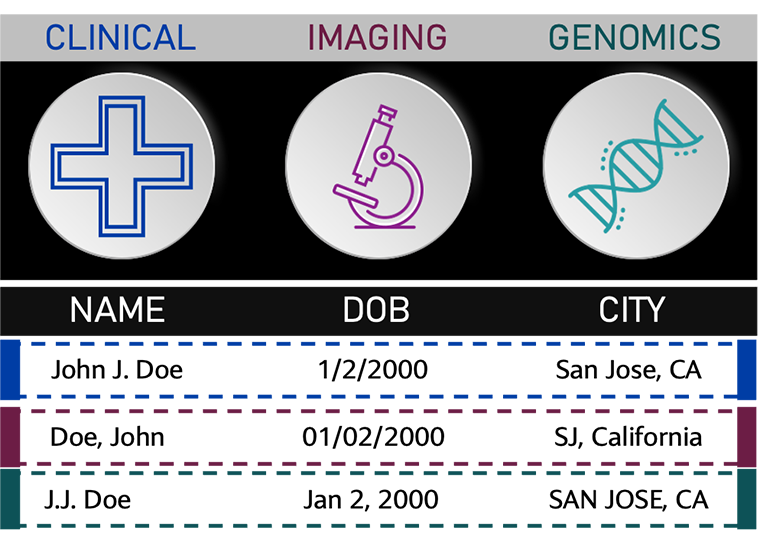 Figure 1. Example of non-standardized data to be encoded for record linkage
Figure 1. Example of non-standardized data to be encoded for record linkage